Classifying Dermatalogical Lesions
Team: Rahil Patel, Sultan Sayezada, Yunni Zhu, Alexander Guo, Chris TurkoCS 4476 - Intro to Computer Vision - Fall 2020
Georgia Institute of Technology
Abstract
Dermatologists in today's medical system are often the ones to diagnose different skin cancers based on visual cues, however, they are not always accurate. Our goal is to instead use a computer vision model in order to recognize and categorize different skin lesions and identify their corresponding skin cancer. For our approach we first used classical computer vision techniques to gather metrics which were specific to our problem and were closely related with how dermatologists typically identify skin lesions. We then used these metrics along with a deep learning model to accurately identify which skin lesions corresponded to each skin cancer. For each lesion, we returned our best prediction for the type of skin cancer based on evidence from our model. Our prediction can then be subsequently used by a dermatologist to recommend the best course of treatment.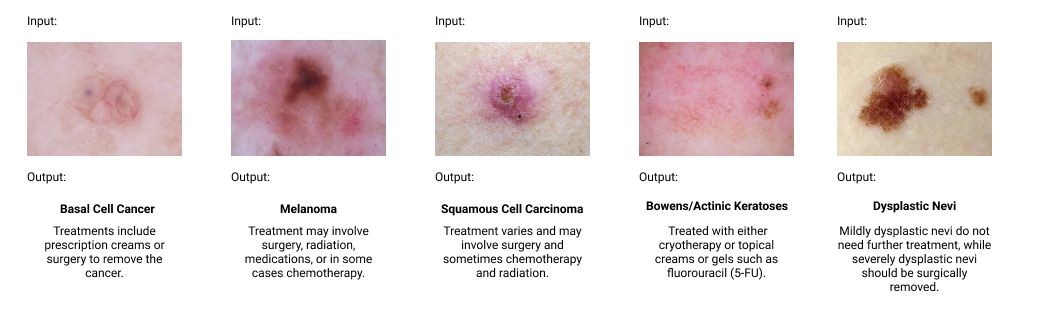
Introduction
The goal of this project is to be able to analyze different pigmented skin lesions in a dermatoscopic image and successfully and accurately classify the type of potential skin cancer with which the lesion is associated. This is an important problem because early detection of skin cancer is imperative for proper treatment. We will be attempting to build a classifier using techniques learned from class as well as extending to modern deep learning based methods. Thus, the input to our system is a dermatoscopic image, and the output would be a corresponding potential skin-cancer which falls within one of seven diagnostic categories.Approach
Our approach incorporates traditional computer vision techniques from class in addition to deep learning based methods. We will need to incorporate edge detection and segmentation/clustering with the methods we learned in Computer Vision to separate the skin lesion from the rest of the image. Some of the main techniques that we will be using include clustering and quantizing RGB pixel values, using the hough transform, calculating shape and circularity, and using gradient information. As an example, the shape of the lesion will give us hints to which category it belongs to. An irregular shape with poorly defined borders can indicate signs of melanoma. Additionally, we can also consider using the texture and chamfer distance to our advantage. Intuitively, we can use all of these values as features and feed them into a basic machine learning algorithm (KNN, MLP) to classify the lesion type. Additionally, we will train various Machine Learning classification models (specifically, K-Nearest Neighbors and Multi-Layer Perceptron classifers) with the data obtained by the previously mentioned Computer Vision techniques in order to classify the images.In addition to these traditional computer vision methods, we have also used a deep convolutional neural network to classify the lesion type. Instead of building our own neural network from scratch, we will fine-tune a pre-existing model such as the ResNet50. Doing this will give us much better accuracy. Another necessary component to this will be experimenting with different hyperparameters, data augmentation, and optimizers. There are many different libraries that allow us to experiment freely such as Pytorch for deep learning, or opencv for other computer vision techniques.
So far, we have faced some difficulty using the classical computer vision techniques to classify skin lesions. There are so many different possible features we can use. Even if we select a group of features, the dataset is very heavily skewed towards several categories. We had to address this by randomly undersampling some of the categories (as you will see below). Also, most of our team members will agree that looking at skin diseases all day is not pleasant at all!
Experiments and Results
We plan on drawing our data from skin cancer datasets found at https://www.kaggle.com/kmader/skin-cancer-mnist-ham10000 and https://www.aad.org/public/diseases/skin-cancer/types. The HAM10000 dataset contains around 10000 dermatoscopic images that are labeled with 7 different categories of skin lesions. We will be using this to help us develop a pipeline using classical computer vision techniques to classify skin lesions. This dataset will also be used to train our deep learning model.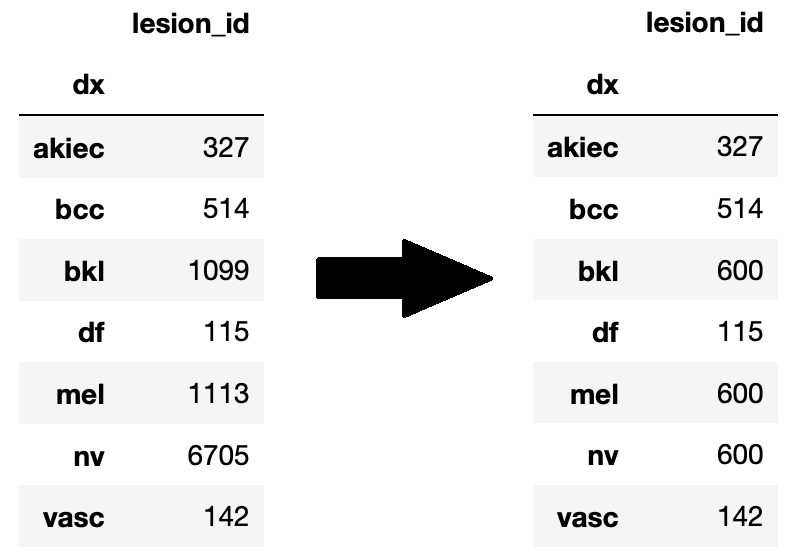
As you can see above, the categories in the HAM10000 are very skewed (for example "nv" which is "Melanocytic nevi"), so we randomly undersampled those categories (n=600) in order to make things more balanced. After doing this, the data was further split 80/10/10 into train/valid/test.
We will evaluate our approaches by looking at how accurately they predict the ground truth in a test set of images. We will also want to calculate both precision and recall, as it is very important to look at false negatives, especially with skin cancer cases. So far, the highest accuracy we've achieved on the test set was 79% using our deep learning approach (finetuning a ResNet50). We achieved this by selecting a good learning rate, and comparing the performance between two different optimizers: SGD, and AdamW (more details in the qualitative results section). In addition to this, we have made headway on using classical computer vision techniques to identify skin lesions. Further experimentation is needed in order to put all of the components together.
We are not surprised by these results, because deep learning is obviously a much more modern and accurate approach. We also expected to run into a little difficulty with the classical computer vision techniques because there are so many different approaches to use! In the section below, we will provide more details about our approach as well as figures and tables.
Qualitative Results
Currently we have two parts to our results section. The first contain our results and metrics which were obtained with classical computer vision techniques while the second will be our results obtained through our deep learning model. As we continue to work on the project, we plan on combining both of these models to obtain more accurate results.Classical Computer Vision:
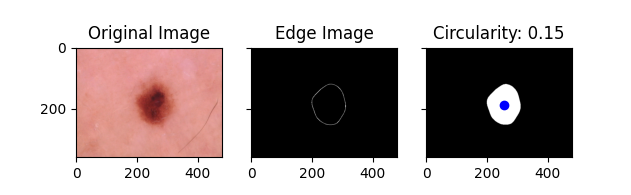
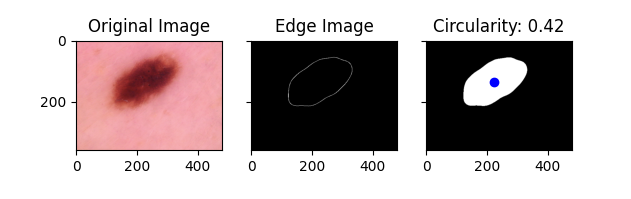
- Circularity Approach One of our metrics obtained using classical computer vision techniques is circularity. In our implementation a value closer to 0 indicates a more circular lesion, while a value closer to 1 indicates a less circular lesion. Circularity is very important in how dermatologists identify basil cell carcinoma. A more circular shape often indicates that a lesion is caused by basil cell carcinoma, while a less circular shape suggests that it is not. Below are some examples of the circularity measures for different lesions:
- Gradient Approach To further utilize the skills we learned in Computer Vision, we examined how the gradient of each image will help to improve our program's accuracy. The gradient output of an image provides us with comprehensive information relating to the texture of each skin condition, which is also crucial to the real world dermatological applications. The gradient measures how quickly the image changes and in which direction it changes. Our program outputs each image's horizontal gradient and vertical gradient. To make better use of this piece of information, we used the Canny edge detector to first find the edges of each skin lesion, then we calculated the sum of the gradient of all the edges to create a data point that will play an essential role in our future implementation.
- Quantize RGB Approach Color is an important component to identifying different types of skin lesions. For example, melanoma is often identified by its medium to dark brown or blue color. Additionally, the difference in color between the skin leison and the color of the skin can indicate different types of skin leisons. Basal cell carcinoma and dermatofibroma are often similar in color to the skin. To determine leison color, we can take the blob area and run a k-means cluster on it. The average cluster centers will be compared to different RGB reference values to determine which possible types of skin lesion is found in an image. Additionally, we can compute a k-means cluster on the entire image (leison + skin) and calculate the standard deviation of the cluster centers. A lower standard deviation (~30) indicates that the leison is a more similar color to the skin, while a higher standard deviation (~60) can indicate a larger contrast in color between the leison and the skin as well as variations of color within the leison.
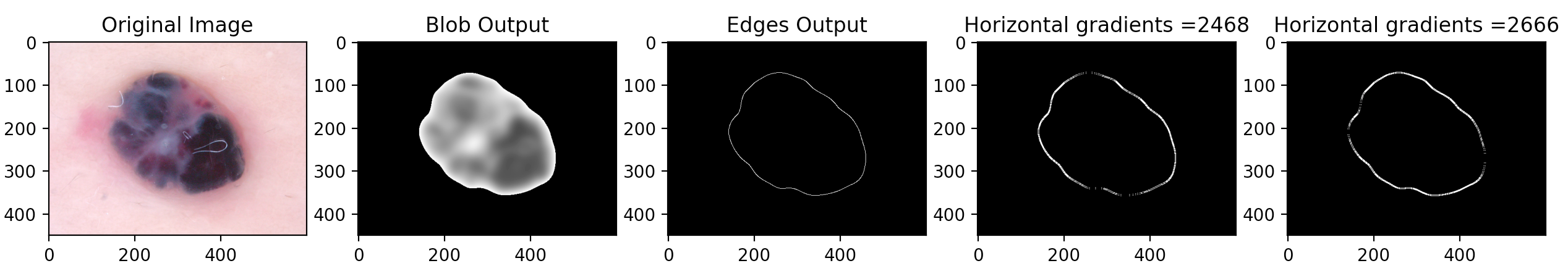
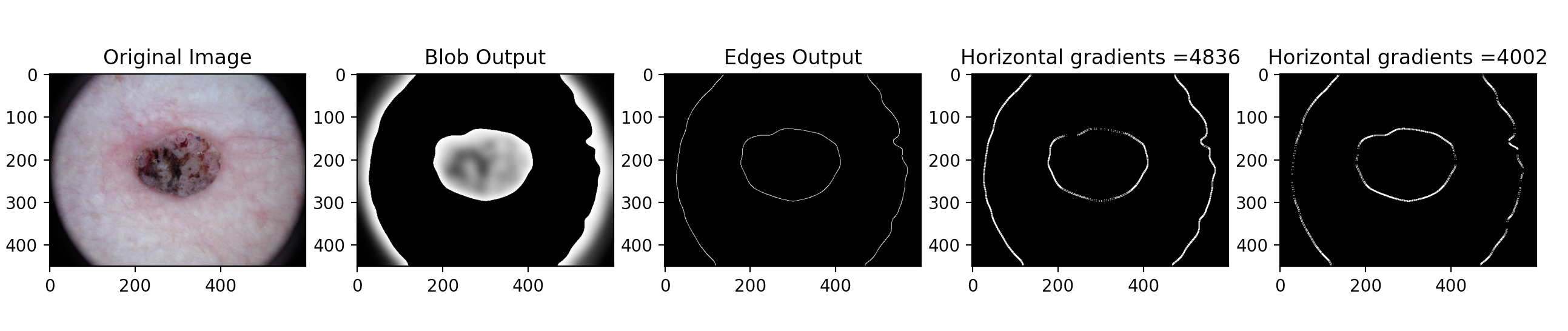
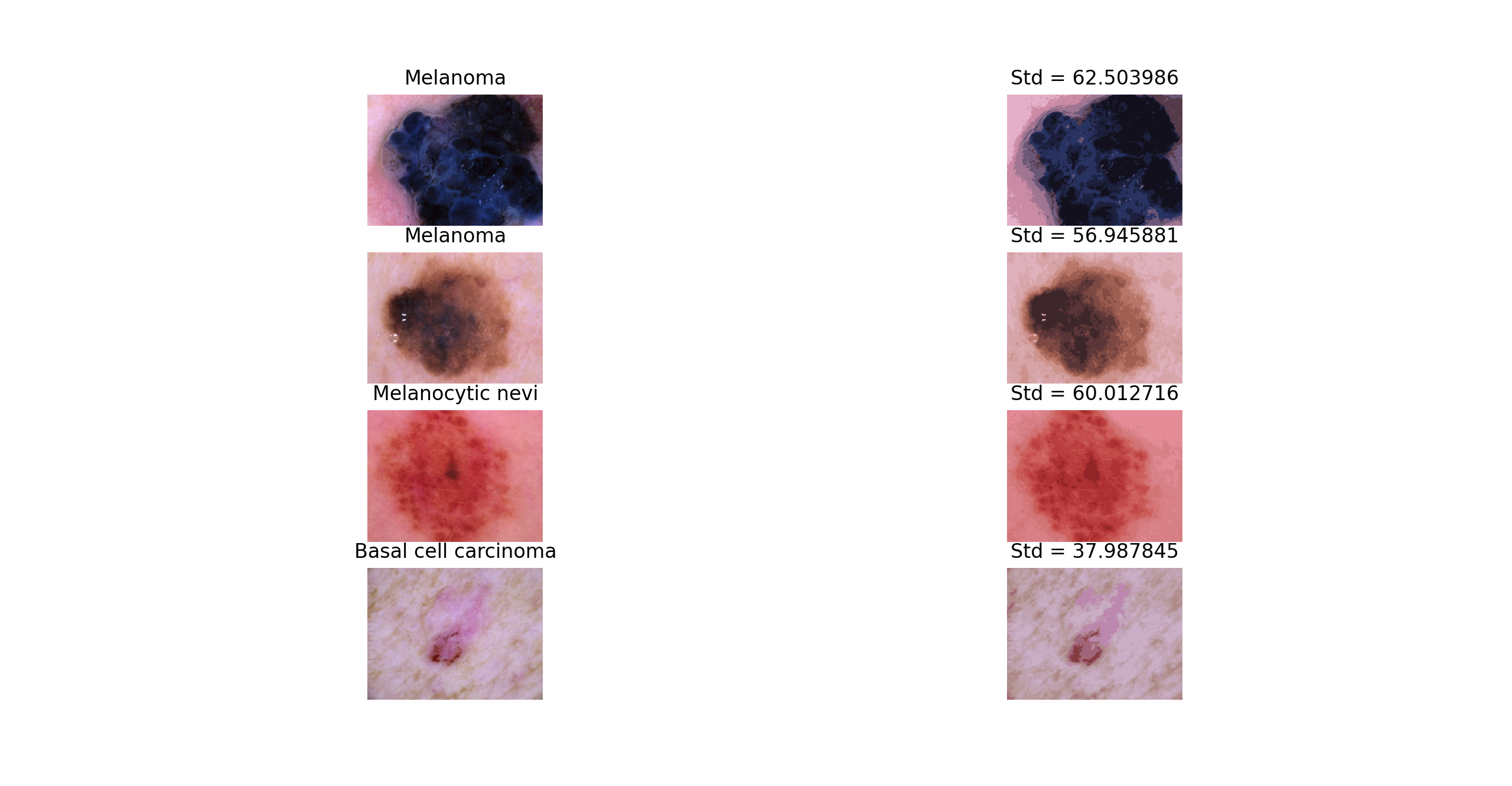
Using Computer Vision with Machine Learning:
After getting the metrics from the previously discussed approaches, such as the circularity and color, we were able to use them as features in a dataset used in the Machine Learning models. The Neural Network, a Multi-layer Perceptron classifer, and a K-Nearest Neighbors model were trained with the data from the Circularity, Gradient, and Quantize RGB functions, with the classifications mapped from string-acronyms to integers. The K-Nearest Neighbors model predicted classifications with a 33.44% accuracy, and the the MLP classifier predicted classifications with a 23.79% accuracy. Although definitely less than optimal, there is definitely promise with these results. Since there were 6 possible classifications, pure guessing would result in a 16.67% accuracy, so our results were much better than pure guessing. We also conducted hyperparameter optimization on the KNN model (varying the algorithm, weight system, and number of neighbors, as shown in the graphs below), in order to increase the accuracy of the KNN model from 24% with base values, to the 33.45% presented here.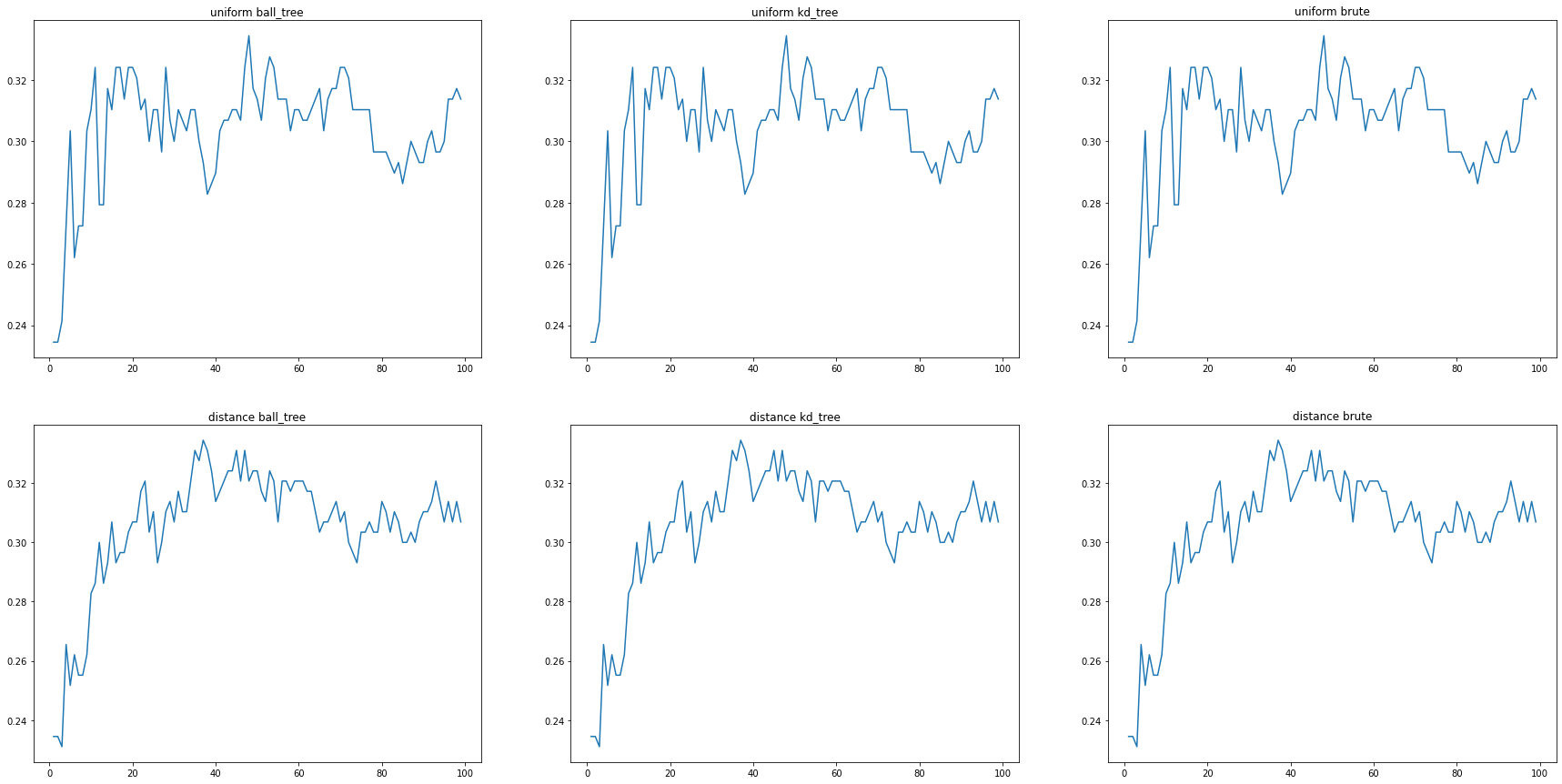
Deep Learning Model:
Through experimentation with our deep learning approach, we were able to achieve 79% accuracy using the AdamW optimizer (learning rate 1e-5) and 78% accuracy using SGD optimizer (learning rate 5e-3). Beneath is the confusion matrix to compare both models. As you would expect, they roughly look the same since there's only a 1% difference in accuracy.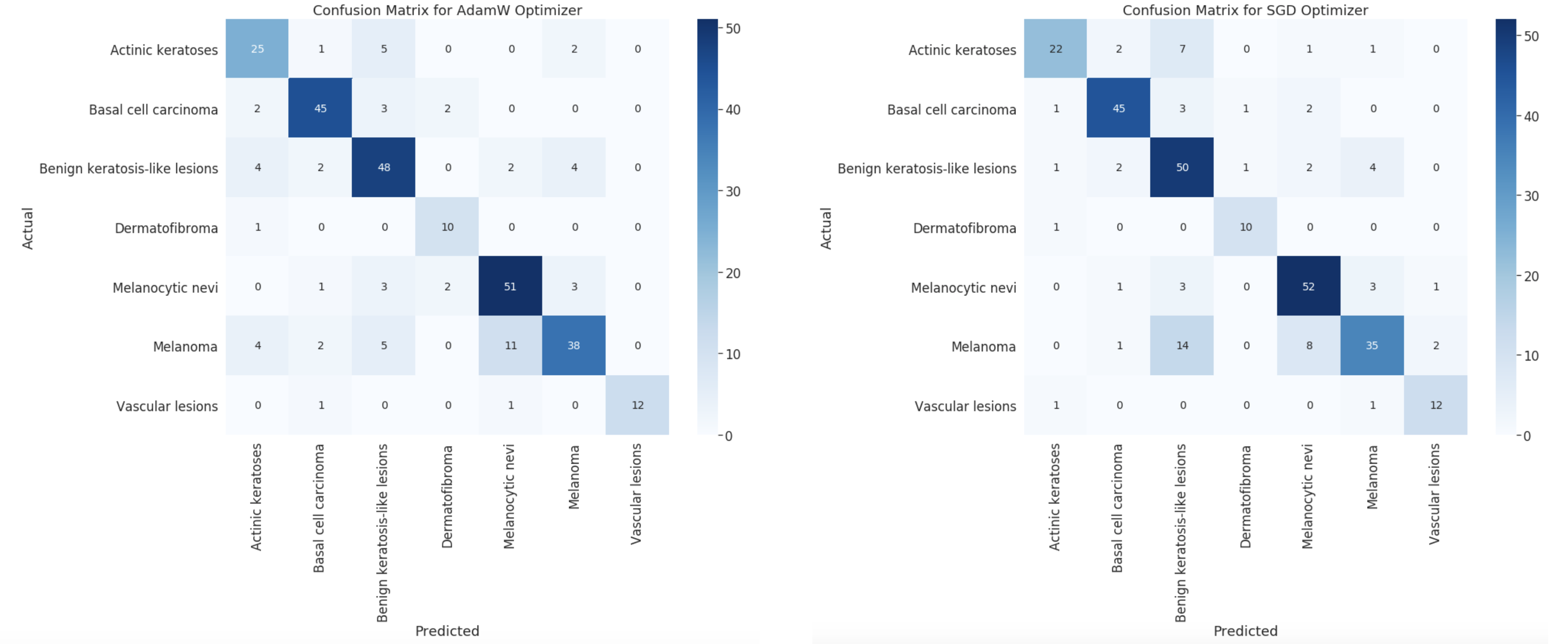
It is no surprise that AdamW performed better, since it is a more advanced optimization technique. In addition to this, we used basic data augmentation such as random affine transformation and flipping during training. We are satisfied with these results, and it is possible to get even higher accuracies if we tested more learning rates, etc. However, since ResNet50 is a huge model and takes a long time to train, it would not be doable in the timespan of this project! Nevertheless, this is clearly a very accurate approach, and it is much better than random guessing (even if there were only 2 categories). We were expecting the deep learning model to perform well, but this honestly exceeded our expectations!
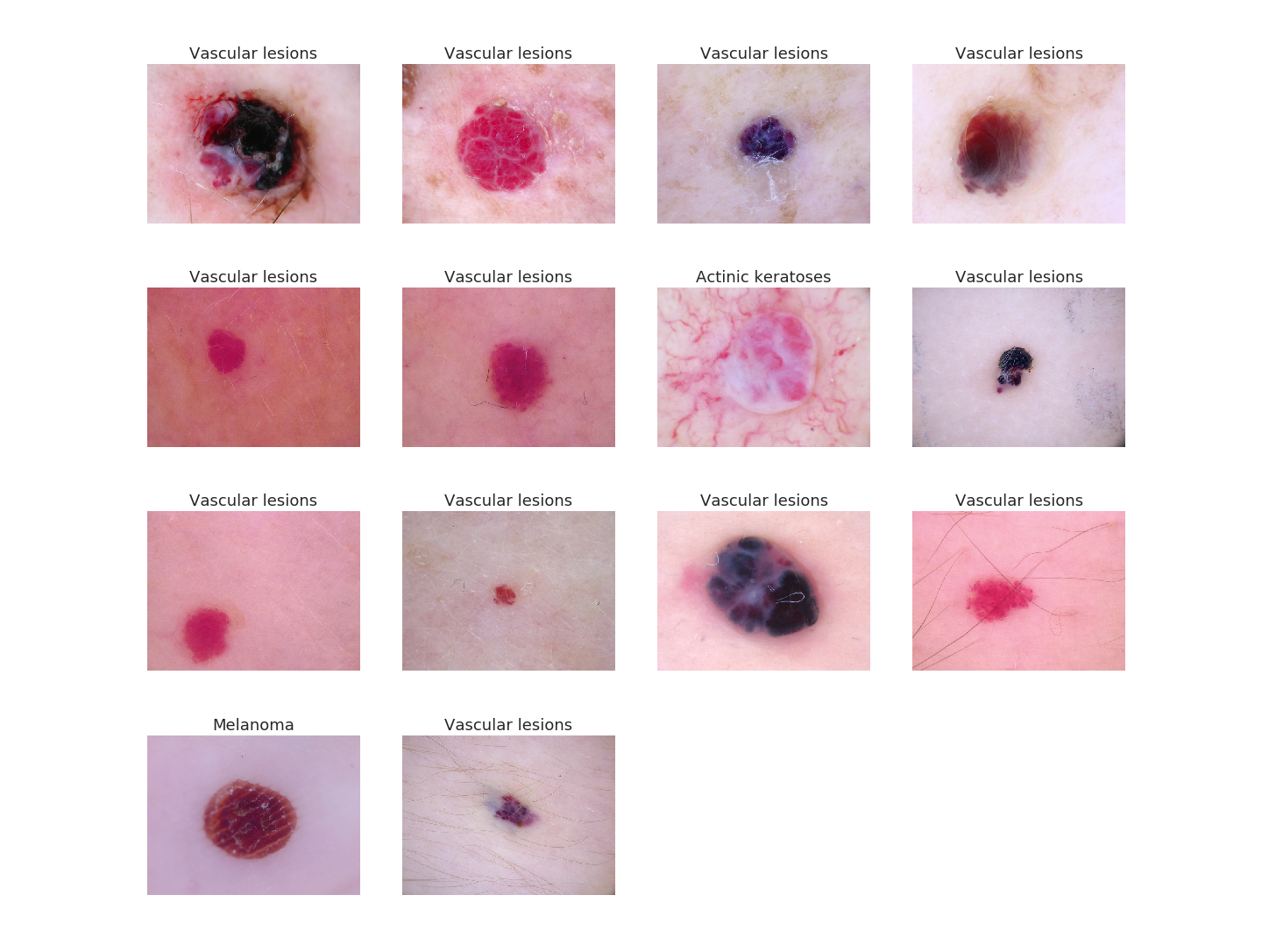
As another qualitative example to show how well our model performs, all of the images below are pictures of vascular lesions. We fed them to our model and ideally we would like the model to label them all as vascular lesions. We did this and the text on top of the image is what the model predicted. As you can see, it was almost able to correctly label all of the images, except for two (row 2, column 3 and row 4, column 1). Can you do better than the model? I don't think so!
Future Work
One thing we can consider doing is using modern data augmentation to enhance the training set. For example, you can use GANs to create new copies of skin lesions. Additionally, there's plenty of more room to experiment with selecting features for the classical computer vision techniques. We can further tune the parameters for the KNN model, and tune the MLP classifier's parameter in order to optimize the accuracy of their results. There are also various other classification methods that we could train the data with, such as Naive Bayes and Random Forest. For the deep learning approach, we can experiment with different pretrained models.Overall, we were able to incorporate both computer vision techniques from class and modern research in order to solve a real world problem. We found out that using the more modern approach yielded a higher accuracy, which is to be expected!
References
Mader, K Scott. “Skin Cancer MNIST: HAM10000.” Kaggle, 20 Sept. 2018, www.kaggle.com/kmader/skin-cancer-mnist-ham10000.“Skin Cancer: Types and Treatment.” American Academy of Dermatology, www.aad.org/public/diseases/skin-cancer/types.